Analysis of miRNA from FFPE
If a technique for expression analysis of genes from FFPE samples can be established, retrospective studies making use of huge volumes of past disease data may be possible, and great contributions to disease and prophylactic treatments could be realized. This idea has attracted attention. However, most FFPE samples are generally stored for a long time, and genes (nucleic acids) may be highly cross-linked, depending on their preparation method. Genes extracted from FFPE samples are highly degraded, especially RNA, and quantitative analysis and evaluation has been considered difficult. However, in fact, RNA is also often degraded during the extraction process. Recently, Toray established a new analysis method as follows:
- Extraction of RNA with good quality
- Determination of whether the extracted RNA has sufficiently high quality for analysis
- Preparation of samples by a method with suppressed detection bias between samples
Regarding (1), we improved on the previous method, in which heat treatment and long-term treatment were necessary, and devised a new method in which extraction is conducted at room temperature in a short time and RNA with a smaller proportion of degraded fractions than that from conventional prescriptions can be extracted (figures below). Regarding (2), we found that the quality can be determined by subjecting RNA to electrophoresis and investigating the presence or absence of degraded fractions and crosslink fractions, as well as their proportions. With this technique, RNA that is very unlikely to show correct analysis results can be excluded beforehand. Regarding (3), miRNAs in particular have short length and are very resistant to degradation. Establishment of quality criteria for appropriate RNA, and adoption of a protocol in which RNA meeting the criteria is not amplified, expand the possibility to obtain valuable information.
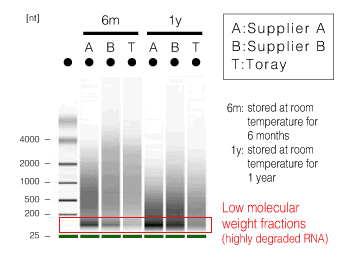
Figure 1. Extraction method and quality of RNA
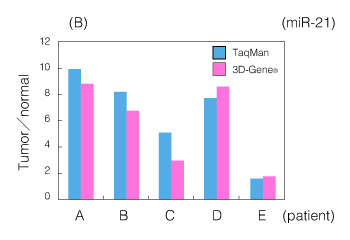
Figure 2. An example of miRNA analysis using an FFPE sample
(B) Comparison with quantitative RT-PCR (miR-21 expression ratio between tumor and normal parts in a gastric cancer FFPE tissue)

