Application notes
Vol.6 Detection specificity for 3D-Gene® miRNA oligo chip
The DNA chip is the tool to detect nucleic acid sequences of the targets DNA /RNA by hybridisation against probes on the chip. Therefore not only sensitivity but also specificity of the hybridization is a very important factor of performance. Thus each manufacturer makes efforts to get high sensitivity and specificity by different solutions. In case of specific hybridization against mRNA, certain specificity is achieved by longer probe length (more than 50mer) or by multiple probes design against single target. However the length of miRNA is only 20~25mer, and makes it difficult to get high specificity only by probe design with sequence specificity. In other words, to improve specificity of hybridization against mature miRNA, not only the probe design is important, but also the high performance DNA microarray substrate and experimental protocol that avoids mismatch hybridization. Because of columnar structure and bead agitation system, 3D-Gene® enables the specific detection for each miRNA. Figure 1, 2 and 3 show the example of detection specificity of let-7 family that was known as the family whose sequence homology among members are very high. The let-7 is one of the first miRNAs founded in C.elegans, and human let-7 family consists of multiple members that show very high homology as in Figure 1 1). Let-7 is called "oncomiR", because it was frequently reported that the expression is changed in cancer tissue or cells 2), and it is also recognized to have an important role in various biological phenomenon. And as shown in figure 2, 3D-Gene® enable to detect each expression with high specificity. And not only synthetic oligoes, but also RNA from biospecimen showed specific detection among family members (Figure 3). As it is shown here, 3D-Gene® detects small difference between sequences, therefore 3D-Gene® can be used as a convenient tool to find the difference of miRNA profiling between samples comprehensively.
References:
- 1) Science 294 (5543): 853-858. 2001
- 2) Mol. Cancer 6:60, 2007

Fig.1 Sequence of human let-7 family
Red characters show mismatch base against let-7a.
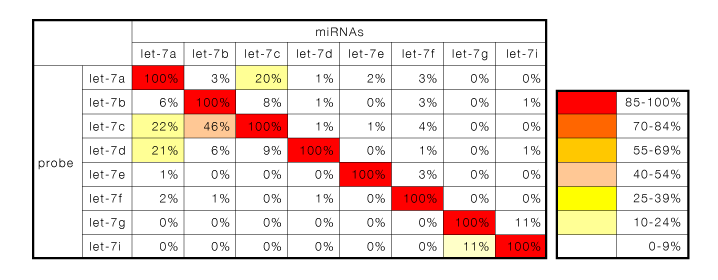
Fig.2 The detection specificity of let-7 family by 3D-Gene®
The same sequences of let-7 family members were synthesized, labelled individually, and detected by 3D-Gene®. The data was normalized as the signal of perfect match sequence between probe and synthesized oligo was considered 100%.
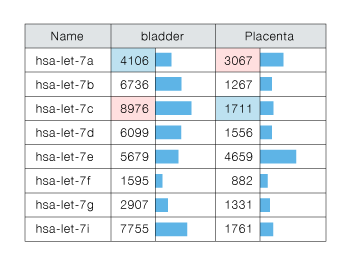
Fig.3 Signal intensity of let-7 family members detected by 3D-Gene®.
The samples are total RNA from human bladder (Ambion, Cat#AM7990) and from human Placenta (Ambion, Cat#AM7950). Bar graph indicates signal intensity of both samples.

