Application notes
Vol.3 Correlation between data obtained by DNA chip and quantitative PCR
DNA chips can detect several hundred to several hundred thousand genes exhaustively from a small amount of specimen in a relatively short time. They are used in many biological studies. The U.S. Food and Drug Administration (FDA) has led the MicroArray Quality Control (MAQC) project1) since 2006 to improve the quality of studies using DNA chips, and examined the validity of detection of gene expression with DNA chips using microarray platforms of 7 companies. The FDA has concluded that detection with DNA chips can be generally standardized regardless of differences in the platform.
Meanwhile, lack in correlation between data obtained from DNA chips and qRT-PCR is still often pointed out as a problem. In fact, correlation with the TaqMan technique is included among evaluation items in the MAQC project, as some genes with poor data correlation are observed on any microarray platform.
This phenomenon may be caused by specificity of the probe design and hybridization conditions among others. We focused on the pretreatment process of mRNA to be detected. That is, RNAs amplified by nonspecific RNA transcription using the T7 primer are currently detected following most DNA chip detection protocols; however, there is no guarantee that all genes are uniformly amplified. This bias may cause the difference in result from data obtained by qRT-PCR. However, mRNAs generally account for only 5% of total RNAs. In a limited amount of specimen, detection of mRNAs is difficult without amplification.
Accordingly, we developed a "non-amplification method" in combination with 3D-Gene®, which can detect mRNAs without amplifying, and its detection results were compared with those of qRT-PCR and amplification methods. As this non-amplification method can detect mRNA reversely transcribed to cDNA directly on the DNA chip, the amplification bias can be avoided. However, as signals are minute, high sensitivity is required. Correlation between the results of detection using 3D-Gene® and data from qRT-PCR in the non-amplification method was improved as expected compared with the amplification method (Fig.1 and 2). The combination of 3D-Gene® and this non-amplification method with high sensitivity can be further applied to experiments with high quantitative performance and can contribute to the development of DNA chips for testing and diagnoses, of which validation is important and detection cost needs to be reduced.
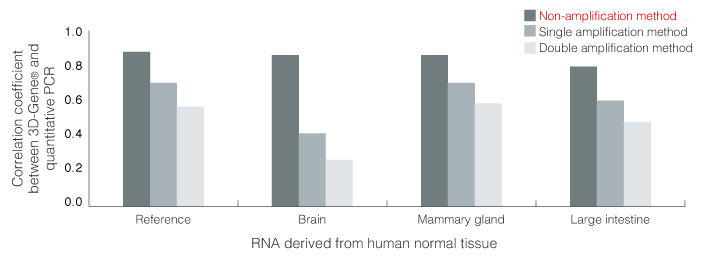
Fig.1 Correlation between data of quantitative RT-PCR and DNA chip
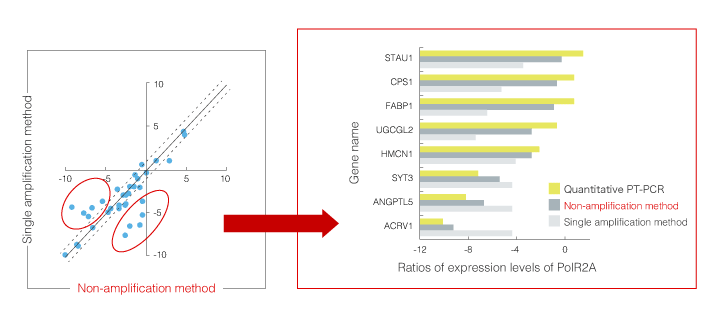
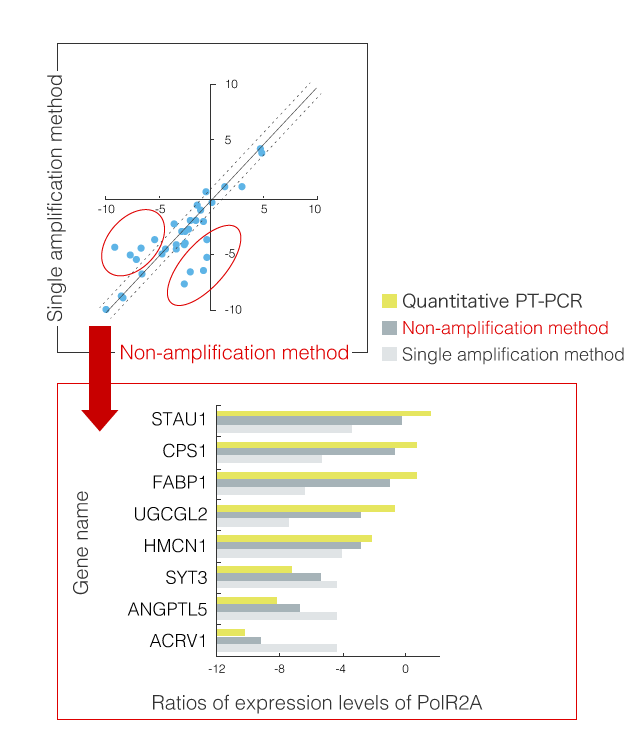
Fig.2 Detection results of non-amplification method and amplification method
References:
- 1) Nature Biotechnology 24(9),2006.

