Application notes
Vol.5 Analysis using miRNA extracted from blood-derived sample
Summary
Cells of higher organisms, including mammals, contain many non-coding RNAs (ncRNAs), which are transcribed but do not code for a protein, unlike conventional RNA (messenger RNA). Among these are single-stranded short RNAs called micro RNAs (miRNAs), which have a length of approximately 20 to 25 nucleotides. It has been elucidated that miRNAs play important roles in the cell differentiation process as well as disease etiology.
In recent years, miRNAs have been shown to exist stably in serum (plasma) and have drawn attention particularly as markers for diagnosis of diseases, evaluation of therapeutic effects, selection of treatment in clinical studies, and others. It has been thought that, as a large amount of RNases exist in serum (plasma), RNAs in blood are degraded in a short time, and miRNA cannot exist in serum. However, recent studies have demonstrated the existence of placenta-derived miRNAs in the serum of a maternal body. Further, studies of miRNAs in blood as a target, such as exhaustive analysis of miRNAs existing in serum, have increased in number dramatically (see References).
In particular, as miRNAs exist in serum (plasma) stably in vesicles released from cells, they have advantages as markers as listed in Table 1. The mRNAs in a vesicle exist only in trace amounts, and analysis with reproducibility is difficult. Therefore, we made use of the high sensitivity feature of the 3D-Gene® DNA chip and established a prescription with which exhaustive miRNA analysis can be performed from a small amount of blood specimen with good reproducibility. The features of this prescription include (1) an extraction method that can obtain high-purity miRNA (high reproducibility and stability), (2) a simple method without complicated extraction operations, and (3) increased number of detected genes and data reproducibility using high sensitivity chips.
- miRNAs can reflect the condition of a lesion (surrounding tissues) and specificity to an injured tissue.
- miRNAs can be easily collected and have good quality as a biomarker because of its stability against freezing/thawing, temperature, acids, and other factors.
- There are fewer types of miRNAs than genes (approximately 1000 types)
- miRNAs have high homology across species, and animal experiment results can be easily extrapolated to humans.
- miRNAs occur early in the protein synthesis and can therefore be targets of drug discovery. Responses are rapid.
- It is suggested that miRNAs are involved in cellular signal transduction via vesicles.
References:
- 1) Chim, S.S. et al.: Clin. Chem., 54: 482-490, 2008
- 2) Gilad, S. Et al.: PloS ONE, 3: e3148, 2008
- 3) Mitchell, P.S. et al.: Proc. Natl. Acad. Sci. USA, 105: 10513-10518, 2008
- 4) Kosaka N, et al.: J Biol Chem., 285(23):17442-52, 2010
- 5) Kosaka N, et al.: Cancer Sci., 101(10):2087-92, 2010
Analysis example
miRNA was prepared from 0.3 mL each of serum from healthy subjects (pooled samples from 10 subjects) and serum from patients with cancer (pooled samples from 6 subjects) in accordance with this prescription, and was subjected to comparison analysis. The results are shown in Fig.1. Fig.2 shows the results of comparison analysis of 0.3 mL each of healthy subject serum from group A (pooled samples from 10 subjects) and group B (pooled samples from 10 subjects) prepared in accordance with this prescription, with 3D-Gene® as control experiment. miRNAs in blood show high correlation between healthy subjects and are found to be expressed stably. On the other hand, miR-X, for which expression is suppressed specifically in cancer patients (Fig.1), has a function of suppressing the expression of oncogene, and is an miRNA known to reduce the expression of two or more types of cancers (lung, ovary, stomach, intestine, and other cancers). Meanwhile, miR-Y, for which expression is increased in patients with cancer, has not been reported to be related to cancers, and its target and function are unknown. We therefore consider that it can be a novel cancer marker candidate.
Thus, the developed analysis prescription of blood miRNA is characterized by miRNA being efficiently extracted from a trace amount of serum (plasma), and a high sensitivity chip is used, reproducibility is high as in gene expression analysis, and quantitative analysis can be performed.
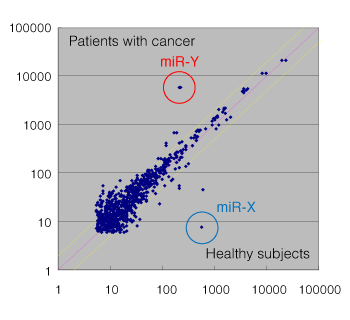 Fig.1 Comparison between healthy subject serum (pooled samples from 10 subjects) and cancer patient serum (pooled samples from 6 subjects)
Fig.1 Comparison between healthy subject serum (pooled samples from 10 subjects) and cancer patient serum (pooled samples from 6 subjects)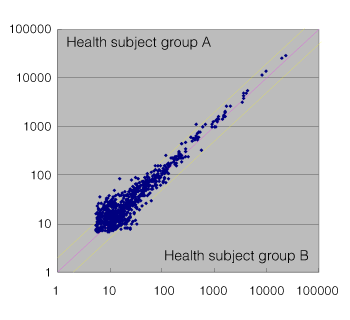 Fig.2 Comparison between healthy subject serum group A (pooled samples from 10 subjects) and healthy subject serum group B (pooled samples from 10 subjects)
Fig.2 Comparison between healthy subject serum group A (pooled samples from 10 subjects) and healthy subject serum group B (pooled samples from 10 subjects)

