Application notes
Vol.2 Gene Expression Analysis Using Entire Yeast Genomic DNA Chips
DNA Chip Research Inc.
Toray Industries Inc.
We present here examples of gene expression analysis in which the 3D-Gene® Yeast Oligo chip 6k was used. This chip is an entire yeast genomic chip composed of a columnar structure. On this chip, probes are mounted for 5,888 genes registered in the Saccharomyces Genome Database (SGD). Gene expression of the budding yeast (Saccharomyces cerevisiae) can be comprehensively analyzed.
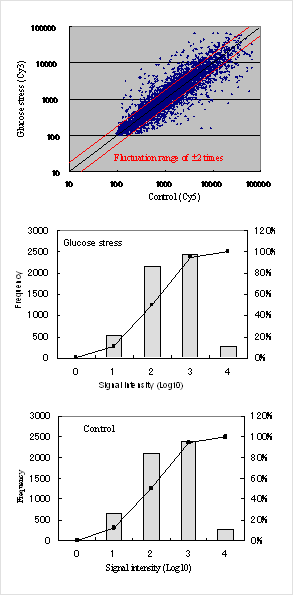 Fig.1 Histograms of yeast gene expression
Fig.1 Histograms of yeast gene expressionFirst, a yeast strain culture in 5 mL YPAD medium was shaken overnight at 30℃ (the preculture). Then 150 mL of YPAD medium was inoculated so that the OD660 became 0.1, and it was shaken and cultured at 30℃ (the main culture). When cell growth corresponded to OD660=0.6, the main culture solution was divided into 10 mL portions. The cells were harvested and washed with sterilized water. The harvested cells from each portion were resuspended in 10 mL of medium without glucose. It was shaken gently for 15-30 min at 30℃. As a control, the cells were resuspended in YPAD medium and then shaken in similar conditions as above. The cells which were harvested from these resuspensions were established as the yeast cells for expression analysis. Total RNA was adjusted using the method of Schmitt et al.1 Expression analysis was performed by competitive hybridization.
Fig.1 shows expression histograms. Our DNA chip has a dynamic range from low to high signals – nearly 4 orders of magnitude. It has the capability to analyze low expression genes with low signal values. Such low signals are difficult to detect using conventional glass DNA chips because of noise.
In this study, we sought to narrow the genes to those with expression specifically induced or inhibited during glucose stress in the exponential growth phase. Under aerobic conditions, glucose is broken down into carbon dioxide and water by metabolic pathways such as the glycolysis pathway, the TCA cycle (citric acid cycle), and the electron transport chain. Then ATP is generated. Many genes which code for enzymes involved in these metabolic pathways have high expression levels. When glucose is depleted, it is known that phosphorylation of glucose is strongly induced by glucokinase (GLK1) and hexokinase (HXK1), and that the expression of a gene group involved in the pentose phosphate pathway and glutamate dehydrogenase (GDH1, GDH3) are strongly induced. Fig. 2 shows the comprehensive analysis results of expression changes of enzymes related to glucose metabolism. While HXK2 expression is inhibited by glucose stress, HXK1 and GLK1 are specifically and strongly induced. These results were consistent with the results of previous studies.2,3 Other than HXK, there are enzyme families, such as PGM and ADH, with highly homologous proteins existing in multiple numbers. In these enzyme families, a gene could be specified that has a specific function. High specificity (selectivity) was confirmed for probes mounted on our chips.
Yeast is one of the organisms which have been studied the longest. There is a wealth of knowledge on the functions of each gene. There are maintained databases with these gene functions and a maintained yeast library4 with factors singly deleted. In the future, functions of ultra-low expression genes need to be comprehensively elucidated using a deletion library and combinations of probes with high specificity and new DNA chip technology, enabling 100-fold higher sensitivity. In addition, databases need to be enhanced. It can be anticipated that then yeast will be the "model animal" of eukaryotic cells which can be used in worldwide research of further wide-ranging fields.
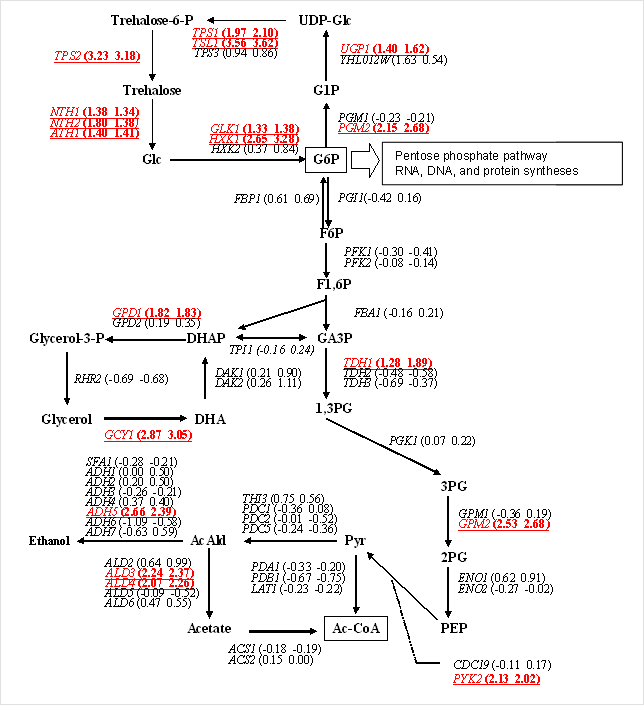
Fig.2 Enzymatic genes involved in the glucose metabolic pathways
[The numbers in parentheses indicate change ratios (log2) in dyeswap experiments.The information in red indicates an expression increase of 2 fold or more.]
References:
- 1) Schmitt, et al. (1990) Nucleic Acids Res, 18:3091-3092.
- 2) Rodriguez A, et al. (2001) Biochem J 355:625-31.
- 3) Lobo Z and Maitra PK (1977) Arch Biochem Biophys 182(2): 639-45
- 4) http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html

