Integrated analysis of miRNA and mRNA
1. Correlation between miRNA and mRNA
MicroRNA (miRNA) is a type of non-coding RNA (RNA that does not encode a protein) with a length of about 22 base. It binds to a complementary sequence in the 3’UTR of mRNA (coding RNA), and thereby regulates the gene expression. It is said that 30-50% of all genes are regulated in this manner. The regulation mechanisms employed by miRNA include transcript degradation and translational suppression. Our method is an analytical procedure focusing on transcript degradation.
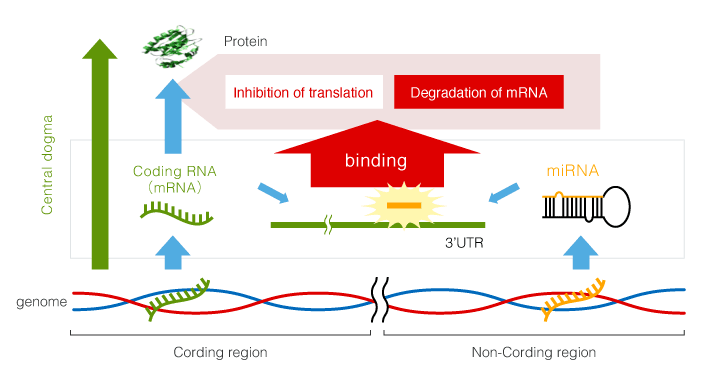
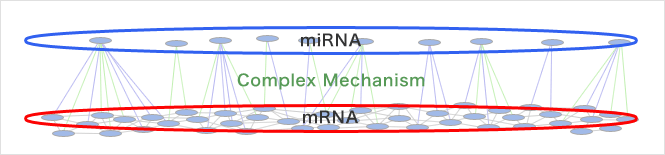
It is considered that an miRNA can regulate gene expression when it has approximately more than 70% homology with the 3’UTR sequence, and thus the regulation mechanism has high flexibility. Therefore, a miRNA regulates many mRNAs, and conversely, a mRNA is regulated by several miRNAs. Correspondence between miRNAs and mRNAs for regulation or non-regulation is not one-to-one: there is a complex correspondence, "many-to-many."
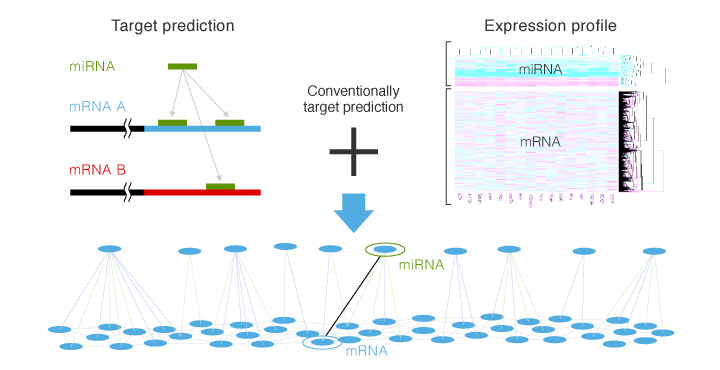
For actual analysis, it is necessary to integrate comprehensive analysis data on miRNAs and mRNAs and data on bioinformatic prediction of target genes to consider by narrowing down their relationship logically and efficiently. This is the concept of integrated analysis.
Examples of websites for target gene prediction
- TargetScan
- http://www.targetscan.org
- PicTar
- http://pictar.mdc-berlin.de/
- mirDB
- http://mirdb.org/miRDB/
- microRNA.org
- http://www.microrna.org/microrna/home.do
- DIANA
- http://diana.imis.athena-innovation.gr/DianaTools/
2. Data analysis for miRNA
In general, differential expressed miRNAs suitable for the purpose of the experimental system are selected.
For example, target variable miRNAs are selected based on the following:
- (1)Expression ratio
- (2)Test between groups
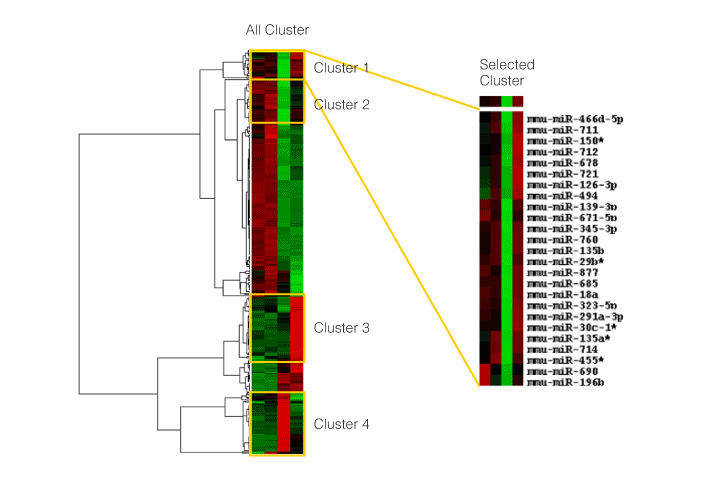
- (3)Cluster analysis
These are general methods used for selecting miRNAs.
If several experimental systems are employed, miRNAs suitable for the purpose are selected by combining them.
(Supplement)
In some cases, location of the selected miRNAs on a gene is confirmed. If some variable miRNAs near (for example, <10 kb) a gene are selected, these miRNAs may be expressed as a cluster under regulation of the same transcription factor and promoter.
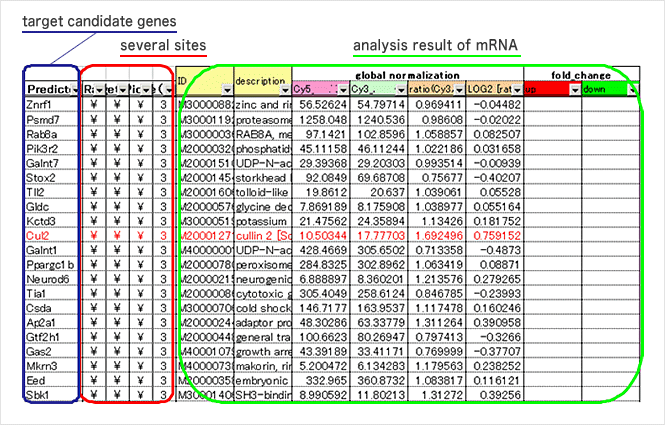
3. Search for target candidates and checking against mRNA analysis data
In regard to miRNAs selected through miRNA data analysis, target candidate genes (mRNAs) are chosen on a website such as TargetScan, Pictar, or miRDB. Genes (mRNAs) that are predicted as targets on several websites are selected* and checked against mRNA array data, and gene candidates that show inverse correlation between expression profiles of miRNA and mRNA are selected.
* This is based on the idea that if the gene is selected by more prediction websites (algorithm), the likelihood may be higher.
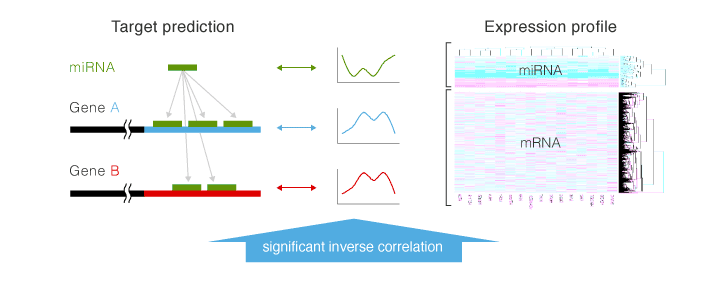
If several data analyses are performed, correlations are compared based on the expression profile of RNAs and miRNAs, and their inverse correlation and relationship with target prediction results are revealed by cluster analysis, etc.
4. GO information for target gene candidates
In addition, GO (Gene Ontology) information is added to mRNA, and functional genes related to the experimental system are selected to extract more meaningful relationship. Inversely, sometimes genes are selected based on GO information, and then miRNAs are reversely determined by the genes, and checked against the analysis data.
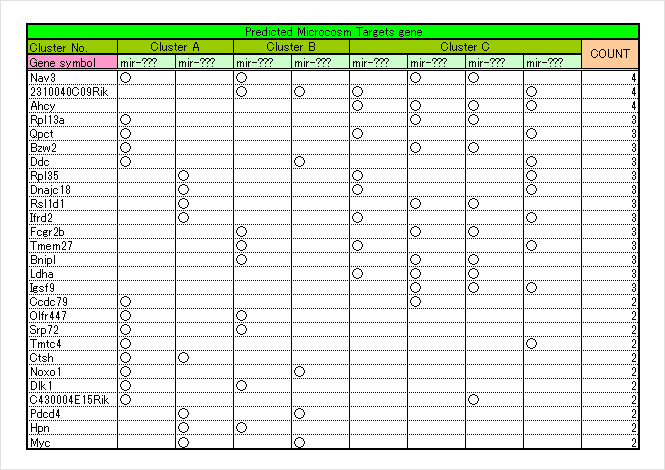
By using combined analysis websites, such as DIANA-mirPath and mirGator, pathway analysis of target genes, collection of GO information, and determination of miRNA targets reversely from the pathway can be performed and the narrowing down of correlation between miRNA-mRNA that is more suitable for the experimental system can be considered.
- DIANA-mirPath
- http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=mirpath/index
- mirGator
- http://genome.ewha.ac.kr/miRGator/miRNAfunction.html
5. Outline of integrated analysis
An example of flow of integrated analysis of miRNA and mRNA is shown below.
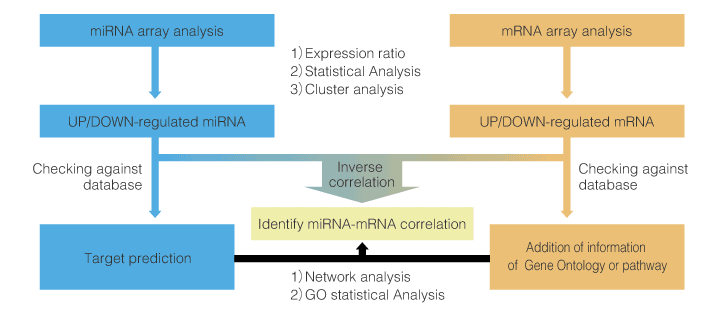
We propose various types of integrated analysis in accordance to the customer's request.

