Performance and advantage of 3D-Gene®
High reproducibility
Since the system has high reproducibility, the data are inherited even if a focused array is created.
The 3D-Gene® Human Digestive Cancer 9k (hereinafter, DC chip) is a focused array created by the extraction of a gene set suitable for digestive system cancer research. It was extracted from a comprehensive type chip, Human Oligo chip 25k (hereinafter 25k chip). The hybrid area is 9000 columns for a DC chip, while it is 25000 columns for a 25k chip, indicating structural differences. The reproducibility of data is assumed to be low between these two chips due to this difference. We used the same sample to examine the differences in data between these chips.
The total RNA was amplified for two types of samples derived from cultured human cells, Samples A and B. Each sample was labeled with Cy3 and Cy5, resulting in labeled samples. The sample adjustment method followed the standard protocol for 3D-Gene®. Sample A-Cy3 and Sample B-Cy5 were applied to a 25k chip. Sample A-Cy5 and Sample B-Cy3 were applied to a DC chip. Thus, hybridization was performed with different combinations (Fig. 1)
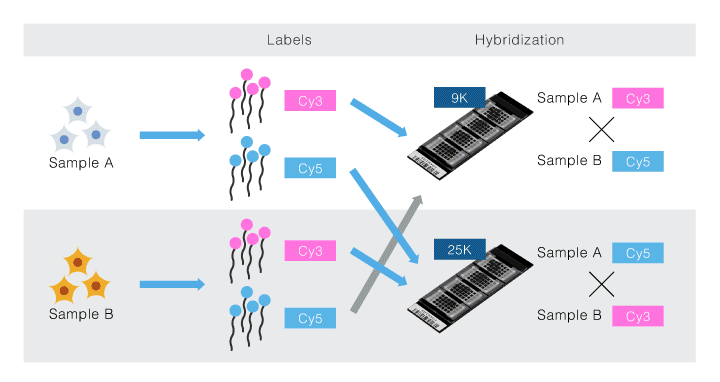
Figure 1. Experiment design
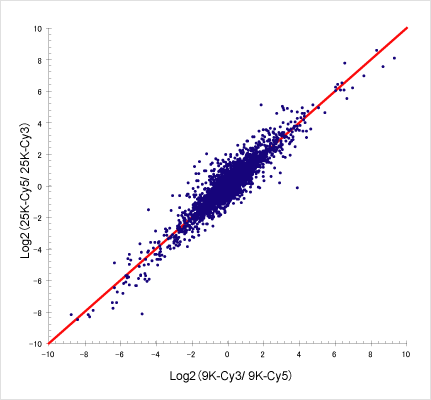
The obtained data were analyzed. The correlation between the microarrays was r=0.98, showing a high correlation despite microarrays being different. Since the 3D-Gene® is a system with high reproducibility, data are good even if the type of microarray changes.
High sensitivity
Even a small amount of mRNA molecules can be detected.
Total RNA (500 ng) in 6 concentrations was added to spike RNA molecules with poly A tails. Total RNA with the spike added was adjusted (Table). As shown in the table of the mole ratios of spike RNA and mRNA, it was assumed that there was a very small amount of RNA molecules. The six types of total RNA with spike added were treated according to the recommended protocol (Fig. 1). Each was hybridized on six 3D-Gene® microarrays. The results are shown in Figure 2. We obtained data with high linearity up to 1 amol addition. Although data variability existed, we confirmed that detection was possible down to 0.1 amol. From the above findings, a 3D-Gene® system can detect RNA molecules in a very minute amount in total RNA.
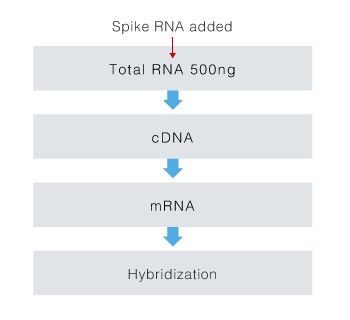
Figure1. Sample processing flow
| Chip ID | added amount(amol) | Spike/mRNA mole ratio |
|---|---|---|
| A | 1000 | 1/1,500 |
| B | 100 | 1/15,000 |
| C | 10 | 1/150,000 |
| D | 1 | 1/1,500,000 |
| E | 0.1 | 1/15,000,000 |
| F | 0.01 | 1/150,000,000 |
Table. Added amount of Spike RNA
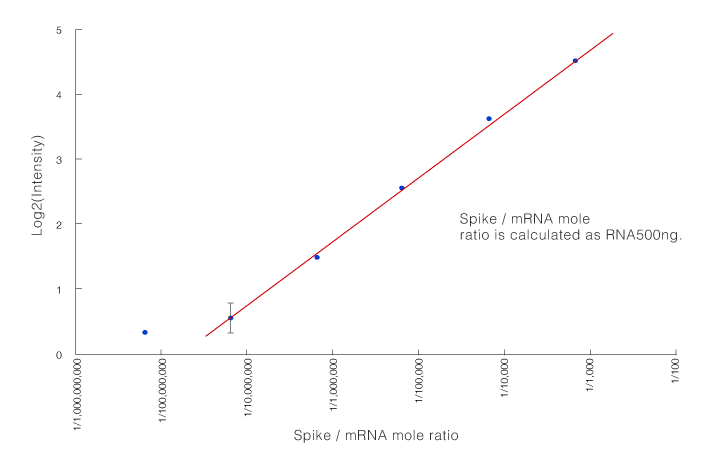
Figure2. Spike/mRNA mole ratio
3D-Gene® is a highly sensitive DNA microarray. Therefore, it is more effective in the detection of low expression genes compared to conventional DNA microarrays. Conventional analysis methods might have missed the changes in expression of genes at very low expression levels. However, the 3D-Gene® can detect such changes in gene expression with high precision. Even if data have been acquired by DNA microarrays from another company, new discovery might be made by analysis using the 3D-Gene®. Please go to analysis examples in the 3D-Gene® application notes.
* For any requested information for 3D-Gene®, please contact us.

